|
|
|
| Alternative polyadenylation: An untapped source for prostate cancer biomarkers and therapeutic targets? |
Akira Kurozumia,Shawn E. Lupolda,b,*( ) )
|
a The James Buchanan Brady Urologic Institute and Department of Urology, Johns Hopkins School of Medicine, Baltimore, MD, USA
b The Department of Oncology, Sidney Kimmel Comprehensive Cancer Center, School of Medicine, Johns Hopkins University, Baltimore, MD, USA |
|
|
|
|
Abstract Objective: To review alternative polyadenylation (APA) as a mechanism of gene regulation and consider potential roles for APA in prostate cancer (PCa) biology and treatment.
Methods: An extensive review of mRNA polyadenylation, APA, and PCa literature was performed. This review article introduces APA and its association with human disease, outlines the mechanisms and components of APA, reviews APA in cancer biology, and considers whether APA may contribute to PCa progression and/or produce novel biomarkers and therapeutic targets for PCa.
Results: Eukaryotic mRNA 3′-end cleavage and polyadenylation play a critical role in gene expression. Most human genes encode more than one polyadenylation signal, and produce more than one transcript isoform, through APA. Polyadenylation can occur throughout the gene body to generate transcripts with differing 3′-termini and coding sequence. Differences in 3′-untranslated regions length can modify post-transcriptional gene regulation by microRNAs and RNA binding proteins, and alter mRNA stability, translation efficiency, and subcellular localization. Distinctive APA patterns are associated with human diseases, tissue origins, and changes in cellular proliferation rate and differentiation state. APA events may therefore generate unique mRNA biomarkers or therapeutic targets in certain cancer types or phenotypic states.
Conclusions: The full extent of cancer-associated and tissue-specific APA events have yet to be defined, and the mechanisms and functional consequences of APA in cancer remain incompletely understood. There is evidence that APA is active in PCa, and that it may be an untapped resource for PCa biomarkers or therapeutic targets.
|
|
Received: 10 November 2020
Available online: 20 October 2021
|
|
Corresponding Authors:
Shawn E. Lupold
E-mail: slupold@jhmi.edu
|
|
|
|
|
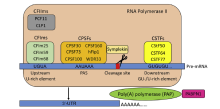
Schematic of the cleavage and polyadenylation complex machinery assembled onto a pre-mRNA polyadenylation site. The CPA complex proteins consist of three major factors: The CPSF complex, the CSTF complex, and the cleavage factor complexes (CFIm and CFIIm), and other factors (Symplekin, PAP, and PABPN1). CPSFs consist of multiple protein subunits (CPSF30, CPSF73, CPSF100, CPSF160, hFip1, and WDR33) that collectively recognize and bind to the PAS specifically. CSTFs consist of three subunits (CSTF50, CSTF64, and CSTF77) that collectively recognize and bind to the GU-/U-rich elements downstream of the PAS. CFIms consist of three subunits (CFIm25, CFIm59, and CFIm68) that collectively recognize and bind to the U-rich elements upstream of the PAS. PAPs and PABPN1 are associated with the addition of untemplated adenosines. CFIIms (PCF11 and CLP1) contribute to transcription termination, and symplekin serves as a scaffold protein of CPA machinery. CPA, cleavage and polyadenylation; CPSF, cleavage and polyadenylation specificity factor; CSTF, cleavage stimulatory factor; PAS, polyadenylation signal; PABPN1, poly(A)-binding protein nuclear 1; PAPs, poly(A) polymerases.
|

|
|
Schematic of CR and UTR APA. APA can be classified as CR-APA and UTR-APA. CR-APA occurs when cleavage and polyadenylation takes place within the coding region or within coding region introns, resulting in truncated protein isoforms. U1snRNP recognizes and blocks early polyadenylation signals to prevent premature cleavage and polyadenylation. UTR-APA occurs when mRNAs encode more than one polyadenylation signal within the 3′-UTR. Proximal polyadenylation generates transcript isoforms with shorter 3′-UTRs and potentially fewer miRNA and RNA binding protein binding sites. Several factors, including NUDT21, CPSF6, and PABPN1 have been reported to inhibit proximal polyadenylation. APA, alternative polyadenylation; CR, coding region; UTR, untranslated region; RBP, RNA binding proteins; PAS, polyadenylation signal.
|
|
|
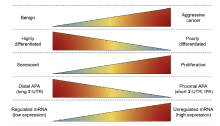
Alternative polyadenylation: An untapped source for prostate cancer biomarkers? Schematic to demonstrate shared relationships between APA and PCa progression. Carcinogenesis and cancer progression are associated with decreasing differentiation state and increasing cellular proliferation. Distal polyadenylation, IPA, and 3′-UTR shortening are similarly associated with proliferation, loss of differentiation, and cancer development. The mechanisms and relationships between APA and cancer remain incompletely defined. We hypothesize that these APA events could create unique transcript isoforms that could serve as diagnostic or prognostic biomarkers for prostate cancer and other urologic malignancies. APA, alternative polyadenylation; IPA, intronic polyadenylation; PCa, prostate cancer; UTR, untranslated region.
|
| [1] |
Miura P, Shenker S, Andreu-Agullo C, Westholm JO, Lai EC. Widespread and extensive lengthening of 3’ UTRs in the mammalian brain. Genome Res 2013; 23:812e25.
|
| [2] |
Ulitsky I, Shkumatava A, Jan CH, Subtelny AO, Koppstein D, Bell GW, et al. Extensive alternative polyadenylation during zebrafish development. Genome Res 2012; 22:2054e66.
|
| [3] |
Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Prolif-erating cells express mRNAs with shortened 3’ untranslated regions and fewer microRNA target sites. Science 2008; 320:1643e7.
|
| [4] |
Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3’ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci U S A 2009; 106:7028e33.
|
| [5] |
Mayr C, Bartel DP. Widespread shortening of 3’UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 2009; 138:673e84.
|
| [6] |
Gruber AJ, Zavolan M. Alternative cleavage and poly-adenylation in health and disease. Nat Rev Genet 2019; 20:599e614.
|
| [7] |
Masamha CP, Wagner EJ. The contribution of alternative pol-yadenylation to the cancer phenotype. Carcinogenesis 2018; 39:2e10.
|
| [8] |
Eckner R, Ellmeier W, Birnstiel ML. Mature mRNA 3’ end for-mation stimulates RNA export from the nucleus. EMBO J 1991; 10:3513e22.
|
| [9] |
Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regula-tion of canonical histone mRNAs: Life without a poly(A) tail. Nat Rev Genet 2008; 9:843e54.
|
| [10] |
Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, et al. A quantitative atlas of polyadenylation in five mammals. Genome Res 2012; 22:1173e83.
|
| [11] |
Mayr C. What are 3’ UTRs doing? Cold Spring Harb Perspect Biol 2019;11:a034728. https://doi.org/10.1101/cshperspect.a034728.
|
| [12] |
Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell 2009; 136:215e33.
|
| [13] |
Kamieniarz-Gdula K, Proudfoot NJ. Transcriptional control by premature termination: A forgotten mechanism. Trends Genet 2019; 35:553e64.
|
| [14] |
Curinha A, Oliveira Braz S, Pereira-Castro I, Cruz A, Moreira A. Implications of polyadenylation in health and disease. Nucleus 2014; 5:508e19.
|
| [15] |
Jenal M, Elkon R, Loayza-Puch F, van Haaften G, Kuhn U, Menzies FM, et al. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell 2012; 149:538e53.
|
| [16] |
Dass B, Tardif S, Park JY, Tian B, Weitlauf HM, Hess RA, et al. Loss of polyadenylation protein tauCstF-64 causes spermato-genic defects and male infertility. Proc Natl Acad Sci U S A 2007; 104:20374e9.
|
| [17] |
Nanavaty V, Abrash EW, Hong C, Park S, Fink EE, Li Z, et al. DNA methylation regulates alternative polyadenylation via CTCF and the cohesin complex. Mol Cell 2020;78:752e64.e6. https://doi.org/10.1016/j.molcel.2020.03.024.
|
| [18] |
Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev 1997; 11:2755e66.
|
| [19] |
Sun Y, Zhang Y, Hamilton K, Manley JL, Shi Y, Walz T, et al. Molecular basis for the recognition of the human AAUAAA polyadenylation signal. Proc Natl Acad Sci U S A 2018;115: E1419e28 https://doi.org/10.1073/pnas.1718723115.
|
| [20] |
Chan SL, Huppertz I, Yao C, Weng L, Moresco JJ, Yates 3rd JR, et al. CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3’ processing. Genes Dev 2014; 28:2370e80.
|
| [21] |
Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res 2000; 10:1001e10.
|
| [22] |
Perez Canadillas JM, Varani G. Recognition of GU-rich poly-adenylation regulatory elements by human CstF-64 protein. EMBO J 2003; 22:2821e30.
|
| [23] |
Beyer K, Dandekar T, Keller W. RNA ligands selected by cleavage stimulation factor contain distinct sequence motifs that function as downstream elements in 3’-end processing of pre-mRNA. J Biol Chem 1997; 272:26769e79.
|
| [24] |
Yang Q, Gilmartin GM, Doublie S. The structure of human cleavage factor I(m) hints at functions beyond UGUA-specific RNA binding: A role in alternative polyadenylation and a po-tential link to 5’ capping and splicing. RNA Biol 2011; 8:748e53.
|
| [25] |
Yang Q, Coseno M, Gilmartin GM, Doublie S. Crystal structure of a human cleavage factor CFI(m)25/CFI(m)68/RNA complex provides an insight into poly(A) site recognition and RNA looping. Structure 2011; 19:368e77.
|
| [26] |
Takagaki Y, Manley JL. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol 2000; 20:1515e25.
|
| [27] |
Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, et al. Polyadenylation factor CPSF-73 is the pre-mRNA 3’-end-processing endonuclease. Nature 2006; 444:953e6.
|
| [28] |
Thuresson AC, Astrom J, Astrom A, Gronvik KO, Virtanen A. Multiple forms of poly(A) polymerases in human cells. Proc Natl Acad Sci U S A 1994; 91:979e83.
|
| [29] |
Lee SH, Singh I, Tisdale S, Abdel-Wahab O, Leslie CS, Mayr C. Widespread intronic polyadenylation inactivates tumour sup-pressor genes in leukaemia. Nature 2018; 561:127e31.
|
| [30] |
Fu Y, Sun Y, Li Y, Li J, Rao X, Chen C, et al. Differential genome-wide profiling of tandem 3’ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res 2011; 21:741e7.
|
| [31] |
Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Canc 2011; 11:558e72.
|
| [32] |
Sanchez G, Bittencourt D, Laud K, Barbier J, Delattre O, Auboeuf D, et al. Alteration of cyclin D1 transcript elongation by a mutated transcription factor up-regulates the oncogenic D1b splice isoform in cancer. Proc Natl Acad Sci U S A 2008; 105:6004e9.
|
| [33] |
Wiestner A, Tehrani M, Chiorazzi M, Wright G, Gibellini F, Nakayama K, et al. Point mutations and genomic deletions in CCND1 create stable truncated cyclin D1 mRNAs that are associated with increased proliferation rate and shorter sur-vival. Blood 2007; 109:4599e606.
|
| [34] |
Wang Q, He G, Hou M, Chen L, Chen S, Xu A, et al. Cell cycle regulation by alternative polyadenylation of CCND1. Sci Rep 2018;8:6824 https://doi.org/10.1038/s41598-018-25141-0.
|
| [35] |
Elliman SJ, Howley BV, Mehta DS, Fearnhead HO, Kemp DM, Barkley LR. Selective repression of the oncogene cyclin D1 by the tumor suppressor miR-206 in cancers. Oncogenesis 2014;3: e113. https://doi.org/10.1038/oncsis.2014.26.
|
| [36] |
Borlado LR, Mendez J. CDC6: From DNA replication to cell cycle checkpoints and oncogenesis. Carcinogenesis 2008; 29:237e43.
|
| [37] |
Akman BH, Can T, Erson-Bensan AE. Estrogen-induced upre-gulation and 3’-UTR shortening of CDC6. Nucleic Acids Res 2012; 40:10679e88.
|
| [38] |
Jin F, Fondell JD. A novel androgen receptor-binding element modulates Cdc6 transcription in prostate cancer cells during cell-cycle progression. Nucleic Acids Res 2009; 37:4826e38.
|
| [39] |
Karanika S, Karantanos T, Li L, Wang J, Park S, Yang G, et al. Targeting DNA damage response in prostate cancer by inhib-iting androgen receptor-CDC6-ATR-Chk1 signaling. Cell Rep 2017; 18:1970e81.
|
| [40] |
Cleynen I, Van de Ven WJ. The HMGA proteins: A myriad of functions (Review). Int J Oncol 2008; 32:289e305.
|
| [41] |
Lee YS, Dutta A. The tumor suppressor microRNA let-7 re-presses the HMGA2 oncogene. Genes Dev 2007; 21:1025e30.
|
| [42] |
Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 2007; 315:1576e9.
|
| [43] |
Hawsawi O, Henderson V, Burton LJ, Dougan J, Nagappan P, Odero-Marah V. High mobility group A2 (HMGA2) promotes EMT via MAPK pathway in prostate cancer. Biochem Biophys Res Commun 2018; 504:196e202.
|
| [44] |
Lovnicki J, Gan Y, Feng T, Li Y, Xie N, Ho CH, et al. LIN28B promotes the development of neuroendocrine prostate can-cer. J Clin Invest 2020; 130:5338e48.
|
| [45] |
Hu R, Dunn TA, Wei S, Isharwal S, Veltri RW, Humphreys E, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Canc Res 2009; 69:16e22.
|
| [46] |
Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 2014; 371:1028e38.
|
| [47] |
Van Etten JL, Nyquist M, Li Y, Yang R, Ho Y, Johnson R, et al. Targeting a single alternative polyadenylation site coordi-nately blocks expression of androgen receptor mRNA splice variants in prostate cancer. Canc Res 2017; 77:5228e35.
|
| [48] |
Masamha CP, Xia Z, Yang J, Albrecht TR, Li M, Shyu AB, et al. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature 2014; 510:412e6.
|
| [49] |
Brumbaugh J, Di Stefano B, Wang X, Borkent M, Forouzmand E, Clowers KJ, et al. Nudt21 controls cell fate by connecting alternative polyadenylation to chromatin signaling. Cell 2018;172:106e20.e21. https://doi.org/10.1016/j.cell.2017.11.023.
|
| [50] |
Jafari Najaf Abadi MH, Shafabakhsh R, Asemi Z, Mirzaei HR, Sahebnasagh R, Mirzaei H, et al. CFIm25 and alternative pol-yadenylation: Con?icting roles in cancer. Canc Lett 2019; 459:112e21.
|
| [51] |
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Canc Dis-cov 2012; 2:401e4.
|
| [52] |
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. https://doi.org/10.1126/scisignal.2004088.
|
| [53] |
Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. UAL-CAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017; 19:649e58.
|
| [54] |
Yang SW, Li L, Connelly JP, Porter SN, Kodali K, Gan H, et al. A cancer-specific ubiquitin ligase drives mRNA alternative pol-yadenylation by ubiquitinating the mRNA 3’ end processing complex. Mol Cell 2020;77:1206e21.e7. https://doi.org/10.1016/j.molcel.2019.12.022.
|
| [55] |
Bai S, He B, Wilson EM. Melanoma antigen gene protein MAGE-11 regulates androgen receptor function by modulating the interdomain interaction. Mol Cell Biol 2005; 25:1238e57.
|
| [56] |
James SR, Cedeno CD, Sharma A, Zhang W, Mohler JL, Odunsi K, et al. DNA methylation and nucleosome occupancy regulate the cancer germline antigen gene MAGEA11. Epige-netics 2013; 8:849e63.
|
| [57] |
Martin G, Gruber AR, Keller W, Zavolan M. Genome-wide analysis of pre-mRNA 3’ end processing reveals a decisive role of human cleavage factor I in the regulation of 3’ UTR length. Cell Rep 2012; 1:753e63.
|
| [58] |
Redis RS, Vela LE, Lu W, Ferreira de Oliveira J, Ivan C, Rodriguez-Aguayo C, et al. Allele-specific reprogramming of cancer metabolism by the long non-coding RNA CCAT2. Mol Cell 2016; 61:520e34.
|
| [59] |
Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet 2007; 39:984e8.
|
| [60] |
Binothman N, Hachim IY, Lebrun JJ, Ali S. CPSF6 is a clinically relevant breast cancer vulnerability target: Role of CPSF6 in breast cancer. EBioMedicine 2017; 21:65e78.
|
| [61] |
Sowalsky AG, Xia Z, Wang L, Zhao H, Chen S, Bubley GJ, et al. Whole transcriptome sequencing reveals extensive unspliced mRNA in metastatic castration-resistant prostate cancer. Mol Canc Res : MCR 2015; 13:98e106.
|
| [62] |
Zheng J, Zhao S, He X, Zheng Z, Bai W, Duan Y, et al. The up-regulation of long non-coding RNA CCAT2 indicates a poor prognosis for prostate cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Biochem Biophys Res Commun 2016; 480:508e14.
|
| [63] |
Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, et al. Genome-wide association study of prostate cancer iden-tifies a second risk locus at 8q24. Nat Genet 2007; 39:645e9.
|
| [64] |
Kuhn U, Gundel M, Knoth A, Kerwitz Y, Rudel S, Wahle E. Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymer-ase and the cleavage and polyadenylation specificity factor. J Biol Chem 2009; 284:22803e14.
|
| [65] |
Hwang HW, Park CY, Goodarzi H, Fak JJ, Mele A, Moore MJ, et al. PAPERCLIP identifies MicroRNA targets and a role of CstF64/64tau in promoting non-canonical poly(A) site usage. Cell Rep 2016; 15:423e35.
|
| [66] |
Ichinose J, Watanabe K, Sano A, Nagase T, Nakajima J, Fukayama M, et al. Alternative polyadenylation is associated with lower expression of PABPN1 and poor prognosis in non-small cell lung cancer. Canc Sci 2014; 105:1135e41.
|
| [67] |
Xiang Y, Ye Y, Lou Y, Yang Y, Cai C, Zhang Z, et al. Compre-hensive characterization of alternative polyadenylation in human cancer. J Natl Cancer Inst 2018; 110:379e89.
|
| [68] |
Stark H, Dube P, Luhrmann R, Kastner B. Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonu-cleoprotein particle. Nature 2001; 409:539e42.
|
| [69] |
Gunderson SI, Beyer K, Martin G, Keller W, Boelens WC, Mattaj LW. The human U1A snRNP protein regulates poly-adenylation via a direct interaction with poly(A) polymerase. Cell 1994; 76:531e41.
|
| [70] |
Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, et al. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 2010; 468:664e8.
|
| [71] |
Berg MG, Singh LN, Younis I, Liu Q, Pinto AM, Kaida D, et al. U1 snRNP determines mRNA length and regulates isoform expression. Cell 2012; 150:53e64.
|
| [72] |
Oh JM, Di C, Venters CC, Guo J, Arai C, So BR, et al. U1 snRNP telescripting regulates a size-function-stratified human genome. Nat Struct Mol Biol 2017; 24:993e9.
|
| [73] |
Oh JM, Venters CC, Di C, Pinto AM, Wan L, Younis I, et al. U1 snRNP regulates cancer cell migration and invasion in vitro. Nat Com-mun 2020;11:1. https://doi.org/10.1038/s41467-019-13993-7.
|
| [74] |
Devany E, Park JY, Murphy MR, Zakusilo G, Baquero J, Zhang X, et al. Intronic cleavage and polyadenylation regulates gene expression during DNA damage response through U1 snRNA. Cell Discov 2016;2:16013. https://doi.org/10.1038/celldisc.2016.13.
|
| [75] |
Dubbury SJ, Boutz PL, Sharp PA. CDK12 regulates DNA repair genes by suppressing intronic polyadenylation. Nature 2018; 564:141e5.
|
| [76] |
Krajewska M, Dries R, Grassetti AV, Dust S, Gao Y, Huang H, et al. CDK12 loss in cancer cells affects DNA damage response genes through premature cleavage and polyadenylation. Nat Commun 2019;10:1757. https://doi.org/10.1038/s41467-019-09703-y.
|
| [77] |
Wu YM, Cieslik M, Lonigro RJ, Vats P, Reimers MA, Cao X, et al. Inactivation of CDK12 delineates a distinct immunogenic class of advanced prostate cancer. Cell 2018;173:1770e82.e14. https://doi.org/10.1016/j.cell.2018.04.034.
|
| [78] |
Lotan TL, Epstein JI. Clinical implications of changing defini-tions within the Gleason grading system. Nat Rev Urol 2010; 7:136e42.
|
| [79] |
Bubendorf L, Sauter G, Moch H, Schmid HP, Gasser TC, Jordan P, et al. Ki67 labelling index: An independent predictor of progression in prostate cancer treated by radical prosta-tectomy. J Pathol 1996; 178:437e41.
|
| [80] |
Fisher G, Yang ZH, Kudahetti S, Moller H, Scardino P, Cuzick J, et al. Prognostic value of Ki-67 for prostate cancer death in a conservatively managed cohort. Br J Canc 2013; 108:271e7.
|
| [1] |
Edward K. Chang,Adam J. Gadzinski,Yaw A. Nyame. Blood and urine biomarkers in prostate cancer: Are we ready for reflex testing in men with an elevated prostate-specific antigen?[J]. Asian Journal of Urology, 2021, 8(4): 343-353. |
| [2] |
Luke P. O’Connor,Shayann Ramedani,Michael Daneshvar,Arvin K. George,Andre Luis Abreu,Giovanni E. Cacciamani,Amir H. Lebastchi. Future perspective of focal therapy for localized prostate cancer[J]. Asian Journal of Urology, 2021, 8(4): 354-361. |
| [3] |
Hamed Ahmadi,Thomas L. Jang,Siamak Daneshmand,Saum Ghodoussipour. Editorial by Bendu K. Konneh, John T. Lafin and Aditya Bagrodia on pp. 341-342 of this issue: MicroRNA-371a-3p as a blood-based biomarker in testis cancer[J]. Asian Journal of Urology, 2021, 8(4): 400-406. |
| [4] |
Wattanachai Ratanapornsompong,Suthep Pacharatakul,Premsant Sangkum,Chareon Leenanupan,Wisoot Kongcharoensombat. Effect of puboprostatic ligament preservation during robotic-assisted laparoscopic radical prostatectomy on early continence: Randomized controlled trial[J]. Asian Journal of Urology, 2021, 8(3): 260-268. |
| [5] |
Zepeng Jia,Yifan Chang,Yan Wang,Jing Li,Min Qu,Feng Zhu,Huan Chen,Bijun Lian,Meimian Hua,Yinghao Sun,Xu Gao. Sustainable functional urethral reconstruction: Maximizing early continence recovery in robotic-assisted radical prostatectomy[J]. Asian Journal of Urology, 2021, 8(1): 126-133. |
| [6] |
Simeng Wen,Yuanjie Niu,Haojie Huang. Posttranslational regulation of androgen dependent and independent androgen receptor activities in prostate cancer[J]. Asian Journal of Urology, 2020, 7(3): 203-218. |
| [7] |
Ieva Eringyte,Joanna N. Zamarbide Losada,Sue M. Powell,Charlotte L. Bevan,Claire E. Fletcher. Coordinated AR and microRNA regulation in prostate cancer[J]. Asian Journal of Urology, 2020, 7(3): 233-250. |
| [8] |
Yezi Zhu,Jun Luo. Regulation of androgen receptor variants in prostate cancer[J]. Asian Journal of Urology, 2020, 7(3): 251-257. |
| [9] |
Ramesh Narayanan. Therapeutic targeting of the androgen receptor (AR) and AR variants in prostate cancer[J]. Asian Journal of Urology, 2020, 7(3): 271-283. |
| [10] |
Yinghao Sun,Liping Xie,Tao Xu,Jørn S. Jakobsen,Weiqing Han,Per S. Sørensen,Xiaofeng Wang. Efficacy and safety of degarelix in patients with prostate cancer: Results from a phase III study in China[J]. Asian Journal of Urology, 2020, 7(3): 301-308. |
| [11] |
Anne Holck Storås,Martin G. Sanda,Olatz Garin,Peter Chang,Dattatraya Patil,Catrina Crociani,Jose Francisco Suarez,Milada Cvancarova,Jon Håvard Loge,Sophie D. Fosså. A prospective study of patient reported urinary incontinence among American, Norwegian and Spanish men 1 year after prostatectomy[J]. Asian Journal of Urology, 2020, 7(2): 161-169. |
| [12] |
Huan Chen,Bijun Lian,Zhenyang Dong,Yan Wang,Min Qu,Feng Zhu,Yinghao Sun,Xu Gao. Experience of one single surgeon with the first 500 robot-assisted laparoscopic prostatectomy cases in mainland China[J]. Asian Journal of Urology, 2020, 7(2): 170-176. |
| [13] |
Kerri Beckmann,Michael O’Callaghan,Andrew Vincent,Penelope Cohen,Martin Borg,David Roder,Sue Evans,Jeremy Millar,Kim Moretti. Extent and predictors of grade upgrading and downgrading in an Australian cohort according to the new prostate cancer grade groupings[J]. Asian Journal of Urology, 2019, 6(4): 321-329. |
| [14] |
Brian T. Hanyok,Mary M. Everist,Lauren E. Howard,Amanda M. De Hoedt,William J. Aronson,Matthew R. Cooperberg,Christopher J. Kane,Christopher L. Amling,Martha K. Terris,Stephen J. Freedland. Practice patterns and outcomes of equivocal bone scans for patients with castration-resistant prostate cancer: Results from SEARCH[J]. Asian Journal of Urology, 2019, 6(3): 242-248. |
| [15] |
Yifan Chang,Xiaojun Lu,Qingliang Zhu,Chuanliang Xu,Yinghao Sun,Shancheng Ren. Single-port transperitoneal robotic-assisted laparoscopic radical prostatectomy (spRALP): Initial experience[J]. Asian Journal of Urology, 2019, 6(3): 294-297. |
|
|
|
|




