|
|
|
| Androgen receptor co-regulation in prostate cancer |
Dhirodatta Senapatia,Sangeeta Kumaria,Hannelore V. Heemersa,b,c,*( ) )
|
a Department of Cancer Biology, Cleveland Clinic, Cleveland, OH, USA
b Department of Urology, Cleveland Clinic, Cleveland, OH, USA
c Department of Hematology/Medical Oncology, Cleveland Clinic, Cleveland, OH, USA |
|
|
|
|
Abstract Prostate cancer (PCa) progression relies on androgen receptor (AR) action. Preventing AR's ligand-activation is the frontline treatment for metastatic PCa. Androgen deprivation therapy (ADT) that inhibits AR ligand-binding initially induces remission but eventually fails, mainly because of adaptive PCa responses that restore AR action. The vast majority of castration-resistant PCa (CRPC) continues to rely on AR activity. Novel therapeutic strategies are being explored that involve targeting other critical AR domains such as those that mediate its constitutively active transactivation function, its DNA binding ability, or its interaction with co-operating transcriptional regulators. Considerable molecular and clinical variability has been found in AR's interaction with its ligands, DNA binding motifs, and its associated coregulators and transcription factors. Here, we review evidence that each of these levels of AR regulation can individually and differentially impact transcription by AR. In addition, we examine emerging insights suggesting that each can also impact the other, and that all three may collaborate to induce gene-specific AR target gene expression, likely via AR allosteric effects. For the purpose of this review, we refer to the modulating influence of these differential and/or interdependent contributions of ligands, cognate DNA-binding motifs and critical regulatory protein interactions on AR's transcriptional output, which may influence the efficiency of the novel PCa therapeutic approaches under consideration, as co-regulation of AR activity.
|
|
Received: 19 April 2019
Available online: 20 July 2020
|
|
Corresponding Authors:
Hannelore V. Heemers
E-mail: heemerh@ccf.org
|
|
|
|
|
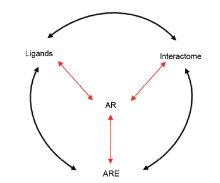
Key determinants of AR transcriptional activity. AR transcriptional output requires ligand activation, binding to cognate DNA binding motifs known as AREs, and interaction with coregulators and secondary transcription factors (interactome). Interactions between these three determinants may fine-tune AR transcription factor function at target genes. AR, androgen receptor; AREs, androgen response elements.
|

| Genes | COSMIC | cBIO | | # | ADT-naïve
n=1 976
% | Barbieri
# | ADT-naïve
n=112
% | Fraser
# | ADT-naïve
n=477
% | TCGA
# | ADT-naïve
n=333
% | ADT-naïve
Armenia
# | + CRPC
n=1 013
% | Abida
# | CRPC
n=444
% | Beltran
# | CRPC
n=114
% | ADT-naïve
n=922
average% | CRPC
n=558
average% | | ABCG5 | 9 | 0.46 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 5 | 0.49 | 6 | 1.35 | 1 | 0.88 | 0.00 | 1.11 | | ABCG8 | 5 | 0.25 | 0 | 0.00 | 0 | 0.00 | 1 | 0.30 | 5 | 0.49 | 4 | 0.90 | 0 | 0.00 | 0.10 | 0.45 | | AKR1C1 | 5 | 0.25 | 0 | 0.00 | 0 | 0.00 | 1 | 0.30 | 3 | 0.30 | 1 | 0.23 | 0 | 0.00 | 0.10 | 0.11 | | AKR1C2 | 1 | 0.05 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 1 | 0.10 | 2 | 0.45 | 0 | 0.00 | 0.00 | 0.23 | | AKR1C3 | 1 | 0.05 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 3 | 0.30 | 1 | 0.23 | 1 | 0.88 | 0.00 | 0.55 | | AKR1C4 | 1 | 0.05 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 3 | 0.30 | 2 | 0.45 | 1 | 0.88 | 0.00 | 0.66 | | CYP11A1 | 8 | 0.40 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 2 | 0.20 | 1 | 0.23 | 0 | 0.00 | 0.00 | 0.11 | | CYP11B1 | 8 | 0.40 | 1 | 0.89 | 0 | 0.00 | 0 | 0.00 | 10 | 0.99 | 8 | 1.80 | 2 | 1.75 | 0.30 | 1.78 | | CYP11B2 | 7 | 0.35 | 0 | 0.00 | 0 | 0.00 | 1 | 0.30 | 8 | 0.79 | 6 | 1.35 | 0 | 0.00 | 0.10 | 0.68 | | CYP17A1 | 1 | 0.05 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 2 | 0.20 | 1 | 0.23 | 0 | 0.00 | 0.00 | 0.11 | | CYP19A1 | 7 | 0.35 | 0 | 0.00 | 2 | 0.42 | 1 | 0.30 | 3 | 0.30 | 1 | 0.23 | 0 | 0.00 | 0.24 | 0.11 | | CYP1A1 | 5 | 0.25 | 1 | 0.89 | 0 | 0.00 | 1 | 0.30 | 5 | 0.49 | 2 | 0.45 | 1 | 0.88 | 0.40 | 0.66 | | CYP1B1 | 4 | 0.20 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 3 | 0.30 | 4 | 0.90 | 0 | 0.00 | 0.00 | 0.45 | | CYP21A2 | 8 | 0.40 | 0 | 0.00 | 0 | 0.00 | 1 | 0.30 | 3 | 0.30 | 2 | 0.45 | 0 | 0.00 | 0.10 | 0.23 | | CYP3A4 | 1 | 0.05 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 3 | 0.30 | 1 | 0.23 | 0 | 0.00 | 0.00 | 0.11 | | CYP3A43 | 2 | 0.10 | 0 | 0.00 | 0 | 0.00 | 2 | 0.60 | 3 | 0.30 | 2 | 0.45 | 0 | 0.00 | 0.20 | 0.23 | | CYP3A5 | 2 | 0.10 | 1 | 0.89 | 1 | 0.21 | 0 | 0.00 | 4 | 0.39 | 1 | 0.23 | 1 | 0.88 | 0.37 | 0.55 | | CYP3A7 | 3 | 0.15 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 2 | 0.20 | 1 | 0.23 | 0 | 0.00 | 0.00 | 0.11 | | HSD17B1 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 1 | 0.10 | 0 | 0.00 | 1 | 0.88 | 0.00 | 0.44 | | HSD17B10 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0.00 | 0.00 | | HSD17B11 | 1 | 0.05 | 0 | 0.00 | 0 | 0.00 | 2 | 0.60 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0.20 | 0.00 | | HSD17B12 | 2 | 0.10 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0.00 | 0.00 | | HSD17B13 | 1 | 0.05 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0.00 | 0.00 | | HSD17B14 | 2 | 0.10 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 2 | 0.20 | 2 | 0.45 | 1 | 0.88 | 0.00 | 0.66 | | HSD17B2 | 1 | 0.05 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 3 | 0.30 | 3 | 0.68 | 0 | 0.00 | 0.00 | 0.34 | | HSD17B3 | 14 | 0.71 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 4 | 0.39 | 2 | 0.45 | 0 | 0.00 | 0.00 | 0.23 | | HSD17B4 | 4 | 0.20 | 0 | 0.00 | 0 | 0.00 | 1 | 0.30 | 5 | 0.49 | 3 | 0.68 | 0 | 0.00 | 0.10 | 0.34 | | HSD17B6 | 2 | 0.10 | 0 | 0.00 | 1 | 0.21 | 0 | 0.00 | 2 | 0.20 | 1 | 0.23 | 1 | 0.88 | 0.07 | 0.55 | | HSD17B7 | 2 | 0.10 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 1 | 0.23 | 0 | 0.00 | 0.00 | 0.11 | | HSD17B8 | 1 | 0.05 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 1 | 0.10 | 0 | 0.00 | 0 | 0.00 | 0.00 | 0.00 | | HSD3B1 | 5 | 0.25 | 0 | 0.00 | 0 | 0.00 | 1 | 0.30 | 2 | 0.20 | 0 | 0.00 | 0 | 0.00 | 0.10 | 0.00 | | HSD3B2 | 1 | 0.05 | 0 | 0.00 | 0 | 0.00 | 1 | 0.30 | 3 | 0.30 | 1 | 0.23 | 0 | 0.00 | 0.10 | 0.11 | | HSD3B7 | 6 | 0.30 | 1 | 0.89 | 0 | 0.00 | 0 | 0.00 | 4 | 0.39 | 2 | 0.45 | 0 | 0.00 | 0.30 | 0.23 | | RDH15 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 1 | 0.10 | 1 | 0.23 | 0 | 0.00 | 0.00 | 0.11 | | RDH16 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 3 | 0.30 | 2 | 0.45 | 0 | 0.00 | 0.00 | 0.23 | | RDH5 | 1 | 0.05 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0.00 | 0.00 | | SHBG | 2 | 0.10 | 0 | 0.00 | 0 | 0.00 | 1 | 0.30 | 1 | 0.10 | 1 | 0.23 | 0 | 0.00 | 0.10 | 0.11 | | SLCO1A2 | 5 | 0.25 | 0 | 0.00 | 1 | 0.21 | 0 | 0.00 | 6 | 0.59 | 2 | 0.45 | 0 | 0.00 | 0.07 | 0.23 | | SLCO1B1 | 6 | 0.30 | 0 | 0.00 | 1 | 0.21 | 1 | 0.30 | 4 | 0.39 | 0 | 0.00 | 0 | 0.00 | 0.17 | 0.00 | | SLCO1B3 | 5 | 0.25 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 2 | 0.20 | 1 | 0.23 | 0 | 0.00 | 0.00 | 0.11 | | SLCO1C1 | 5 | 0.25 | 0 | 0.00 | 0 | 0.00 | 1 | 0.30 | 4 | 0.39 | 1 | 0.23 | 0 | 0.00 | 0.10 | 0.11 | | SLCO2A1 | 5 | 0.25 | 1 | 0.89 | 0 | 0.00 | 1 | 0.30 | 4 | 0.39 | 5 | 1.13 | 0 | 0.00 | 0.40 | 0.56 | | SLCO2B1 | 7 | 0.35 | 0 | 0.00 | 0 | 0.00 | 1 | 0.30 | 8 | 0.79 | 3 | 0.68 | 6 | 5.26 | 0.10 | 2.97 | | SLCO3A1 | 18 | 0.91 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 9 | 0.89 | 3 | 0.68 | 1 | 0.88 | 0.00 | 0.78 | | SLCO4A1 | 9 | 0.46 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 5 | 0.49 | 3 | 0.68 | 2 | 1.75 | 0.00 | 1.22 | | SLCO4C1 | 8 | 0.40 | 4 | 3.57 | 1 | 0.21 | 0 | 0.00 | 12 | 1.18 | 5 | 1.13 | 0 | 0.00 | 1.26 | 0.56 | | SLCO5A1 | 11 | 0.56 | 0 | 0.00 | 0 | 0.00 | 1 | 0.30 | 5 | 0.49 | 5 | 1.13 | 0 | 0.00 | 0.10 | 0.56 | | SLCO6A1 | 12 | 0.61 | 1 | 0.89 | 0 | 0.00 | 1 | 0.30 | 14 | 1.38 | 9 | 2.03 | 0 | 0.00 | 0.40 | 1.01 | | SRD5A1 | 1 | 0.05 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0.00 | 0.00 | | SRD5A2 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 0.00 | 0.00 | | SRD5A3 | 3 | 0.15 | 0 | 0.00 | 1 | 0.21 | 0 | 0.00 | 3 | 0.30 | 3 | 0.68 | 0 | 0.00 | 0.07 | 0.34 | | STAR | 3 | 0.15 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 2 | 0.20 | 2 | 0.45 | 1 | 0.88 | 0.00 | 0.66 | | UGT2B15 | 2 | 0.10 | 0 | 0.00 | 0 | 0.00 | 3 | 0.90 | 3 | 0.30 | 0 | 0.00 | 0 | 0.00 | 0.30 | 0.00 | | UGT2B17 | 3 | 0.15 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 4 | 0.39 | 0 | 0.00 | 0 | 0.00 | 0.00 | 0.00 | | UGT2B7 | 2 | 0.10 | 0 | 0.00 | 2 | 0.42 | 1 | 0.30 | 7 | 0.69 | 1 | 0.23 | 1 | 0.88 | 0.24 | 0.55 |
|
|
Somatic mutations in steroidogenic genes in clinical PCa tissues. COSMIC and cBIO databases were queried for somatic mutations in genes that encode protein involved in the synthesis, metabolism, and transport of androgens [28]. The COSMIC database was queried via the Cancer browser, selecting for “prostate cancer” and “carcinoma” histology, and retrieving only data from genome-wide sequencing efforts. For cBIO analysis, datasets analyses are marked by first author of study that was examined.
|

| Genes | COSMIC | cBIO | | # | ADT-naïve
n=1 976
% | Barbieri
# | ADT-naïve
n=112
% | Fraser
# | ADT-naïve
n=477
% | TCGA
# | ADT-naïve
n=333
% | ADT-naïve
Armenia
# | + CRPC
n=1 013
% | Abida
# | CRPC
n=444
% | Beltran
# | CRPC
n=114
% | ADT-naïve
n=922
average% | CRPC
n=558
average% | | NCOA1 | 10 | 0.51 | 0 | 0.00 | 0 | 0.00 | 2 | 0.60 | 6 | 0.59 | 2 | 0.45 | 0 | 0.00 | 0.20 | 0.23 | | NCOA2 | 9 | 0.46 | 2 | 1.79 | 1 | 0.21 | 1 | 0.30 | 8 | 0.79 | 4 | 0.90 | 0 | 0.00 | 0.77 | 0.45 | | NCOA3 | 11 | 0.56 | 1 | 0.89 | 3 | 0.63 | 2 | 0.60 | 6 | 0.59 | 2 | 0.45 | 1 | 0.88 | 0.71 | 0.66 | | NCOA4 | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 | 2 | 0.60 | 3 | 0.30 | 1 | 0.23 | 0 | 0.00 | 0.20 | 0.11 | | NCOA6 | 17 | 0.86 | 1 | 0.89 | 1 | 0.21 | 2 | 0.60 | 10 | 0.99 | 4 | 0.90 | 0 | 0.00 | 0.57 | 0.45 | | NCOR1 | 19 | 0.96 | 2 | 1.79 | 4 | 0.84 | 6 | 1.80 | 26 | 2.57 | 13 | 2.93 | 2 | 1.75 | 1.48 | 2.34 | | NCOR2 | 28 | 1.42 | 1 | 0.89 | 1 | 0.21 | 0 | 0.00 | 23 | 2.27 | 14 | 3.15 | 1 | 0.88 | 0.37 | 2.02 |
|
|
Somatic mutations in AR-associated NCoA and NCoR coregulators in clinical PCa. COSMIC and cBio databases were queried for somatic mutations in genes that encode AR-associated NCoAs and NCoRs (28). The COSMIC database was queried via the Cancer browser, selecting for “prostate cancer” and “carcinoma” histology, and retrieving only data from genome-wide sequencing efforts. For cBIO analysis, datasets analyses are marked by first author of study that was examined.
|
|
|
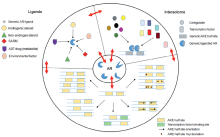
Cooperativity and interdependence between key determinants of AR transcriptional activity are consistent with allosteric regulation of AR action. Schematic representation of variability in AR ligands, AR interactome, and ARE(s) and their influence on one another (outer circle) and their convergence on allosteric regulation of AR, which may exist as different forms (inner circle). ADT, androgen deprivation therapy; AR, androgen receptor; ARE, androgen response element; SARM, selective androgen receptor modulators.
|
| [1] |
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019; 69:7-34.
|
| [2] |
Dai C, Heemers H, Sharifi N. Androgen signaling in prostate cancer. Cold Spring Harb Perspect Med 2017; 7. https://doi.org/10.1101/cshperspect.a030452.
doi: 10.1101/cshperspect.a025528
pmid: 28320828
|
| [3] |
Denmeade SR, Isaacs JT. A history of prostate cancer treatment. Nat Rev Cancer 2002; 2:389-96.
|
| [4] |
Knudsen KE, Penning TM. Partners in crime: deregulation of AR activity and androgen synthesis in prostate cancer. Trends Endocrinol Metab 2010; 21:315-24.
|
| [5] |
Knudsen KE, Scher HI. Starving the addiction: new opportunities for durable suppression of AR signaling in prostate cancer. Clin Cancer Res 2009; 15:4792-8.
|
| [6] |
Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer 2015; 15:701-11.
|
| [7] |
Smith MR, Saad F, Chowdhury S, Oudard S, Hadaschik BA, Graff JN, et al. Apalutamide treatment and metastasis-free survival in prostate cancer. N Engl J Med 2018; 378:1408-18.
|
| [8] |
Attard G, Borre M, Gurney H, Loriot Y, Andresen-Daniil C, Kalleda R, et al. Abiraterone alone or in combination with enzalutamide in metastatic castration-resistant prostate cancer with rising prostate-specific antigen during enzalutamide treatment. J Clin Oncol 2018; 36:2639-46.
|
| [9] |
Davies AH, Beltran H, Zoubeidi A. Cellular plasticity and the neuroendocrine phenotype in prostate cancer. Nat Rev Urol 2018; 15:271-86.
|
| [10] |
Andersen RJ, Mawji NR, Wang J, Wang G, Haile S, Myung JK, et al. Regression of castrate-recurrent prostate cancer by a small-molecule inhibitor of the amino-terminus domain of the androgen receptor. Cancer Cell 2010; 17:535-46.
|
| [11] |
Dalal K, Roshan-Moniri M, Sharma A, Li H, Ban F, Hassona MD, et al. Selectively targeting the DNA-binding domain of the androgen receptor as a prospective therapy for prostate cancer. J Biol Chem 2014; 289:26417-29.
|
| [12] |
Kumari S, Senapati D, Heemers HV. Rationale for the development of alternative forms of androgen deprivation therapy. Endocr Relat Cancer 2017; 24:R275-95. https://doi.org/10.1530/ERC-17-0121.
|
| [13] |
Armstrong CM, Gao AC. Current strategies for targeting the activity of androgen receptor variants. Asian J Urol 2019; 6:42-9.
|
| [14] |
Liu C, Armstrong CM, Lou W, Lombard AP, Cucchiara V, Gu X, et al. Niclosamide and bicalutamide combination treatment overcomes enzalutamide- and bicalutamide-resistant prostate cancer. Mol Cancer Ther 2017; 16:1521-30.
|
| [15] |
Heemers HV, Tindall DJ. Androgen receptor (AR) coregulators: a diversity of functions converging on and regulating the AR transcriptional complex. Endocr Rev 2007; 28:778-808.
|
| [16] |
Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr Rev 2004; 25:276-308.
|
| [17] |
Deslypere JP, Young M, Wilson JD, McPhaul MJ. Testosterone and 5 alpha-dihydrotestosterone interact differently with the androgen receptor to enhance transcription of the MMTV-CAT reporter gene. Mol Cell Endocrinol 1992; 88:15-22.
|
| [18] |
Wilson EM, French FS. Binding properties of androgen receptors. Evidence for identical receptors in rat testis, epididymis, and prostate. J Biol Chem 1976; 251:5620-9.
|
| [19] |
Locke JA, Guns ES, Lubik AA, Adomat HH, Hendy SC, Wood CA, et al. Androgen levels increase by intratumoral de novo steroidogenesis during progression of castration-resistant prostate cancer. Cancer Res 2008; 68:6407-15.
|
| [20] |
Fang H, Tong W, Branham WS, Moland CL, Dial SL, Hong H, et al. Study of 202 natural, synthetic, and environmental chemicals for binding to the androgen receptor. Chem Res Toxicol 2003; 16:1338-58.
|
| [21] |
Cai C, Chen S, Ng P, Bubley GJ, Nelson PS, Mostaghel EA, et al. Intratumoral de novo steroid synthesis activates androgen receptor in castration-resistant prostate cancer and is upregulated by treatment with CYP17A1 inhibitors. Cancer Res 2011; 71:6503-13.
|
| [22] |
Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 2009; 324:787-90.
|
| [23] |
Norris JD, Ellison SJ, Baker JG, Stagg DB, Wardell SE, Park S, et al. Androgen receptor antagonism drives cytochrome P450 17A1 inhibitor efficacy in prostate cancer. J Clin Investig 2017; 127:2326-38.
|
| [24] |
Li Z, Alyamani M, Li J, Rogacki K, Abazeed M, Upadhyay SK, et al. Redirecting abiraterone metabolism to fine-tune prostate cancer anti-androgen therapy. Nature 2016; 533:547-51.
|
| [25] |
Nguyen PL, Alibhai SM, Basaria S, D’Amico AV, Kantoff PW, Keating NL, et al. Adverse effects of androgen deprivation therapy and strategies to mitigate them. Eur Urol 2015; 67:825-36.
|
| [26] |
Pihlajamaa P, Sahu B, Janne OA. Determinants of receptorand tissue-specific actions in androgen signaling. Endocr Rev 2015; 36:357-84.
|
| [27] |
Chisamore MJ, Gentile MA, Dillon GM, Baran M, Gambone C, Riley S, et al. A novel selective androgen receptor modulator (SARM) MK-4541 exerts anti-androgenic activity in the prostate cancer xenograft R-3327G and anabolic activity on skeletal muscle mass & function in castrated mice. J Steroid Biochem Mol Biol 2016; 163:88-97.
|
| [28] |
Cancer Genome Atlas Research Network. Electronic address scmo, cancer genome atlas research N. The molecular taxonomy of primary prostate cancer. Cell 2015; 163:1011-25.
|
| [29] |
Kumar A, Coleman I, Morrissey C, Zhang X, True LD, Gulati R, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med 2016; 22:369-78.
|
| [30] |
Hettel D, Sharifi N. HSD3B1 status as a biomarker of androgen deprivation resistance and implications for prostate cancer. Nat Rev Urol 2018; 15:191-6.
|
| [31] |
Mitsiades N, Sung CC, Schultz N, Danila DC, He B, Eedunuri VK, et al. Distinct patterns of dysregulated expression of enzymes involved in androgen synthesis and metabolism in metastatic prostate cancer tumors. Cancer Res 2012; 72:6142-52.
|
| [32] |
Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castrationresistant prostate cancer. Nature 2012; 487:239-43.
|
| [33] |
DePriest AD, Fiandalo MV, Schlanger S, Heemers F, Mohler JL, Liu S, et al. Regulators of Androgen Action Resource: a one-stop shop for the comprehensive study of androgen receptor action. Database (Oxford). 2016; 2016:bav125. https://doi.org/10.1093/database/bav125.
|
| [34] |
Cato L, de Tribolet-Hardy J, Lee I, Rottenberg JT, Coleman I, Melchers D, et al. ARv7 represses tumor-suppressor genes in castration-resistant prostate cancer. Cancer Cell 2019; 35:401-13.
|
| [35] |
Wang Q, Li W, Liu XS, Carroll JS, Janne OA, Keeton EK, et al. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell 2007; 27:380-92.
|
| [36] |
Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell 2009; 138:245-56.
|
| [37] |
Chen Z, Lan X, Thomas-Ahner JM, Wu D, Liu X, Ye Z, et al. Agonist and antagonist switch DNA motifs recognized by human androgen receptor in prostate cancer. EMBO J 2015; 34:502-16.
|
| [38] |
Chen Z, Wu D, Thomas-Ahner JM, Lu C, Zhao P, Zhang Q, et al. Diverse AR-V7 cistromes in castration-resistant prostate cancer are governed by HoxB13. Proc Natl Acad Sci U S A 2018; 115:6810-5.
|
| [39] |
Massie CE, Lynch A, Ramos-Montoya A, Boren J, Stark R, Fazli L, et al. The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J 2011; 30:2719-33.
|
| [40] |
Chan SC, Selth LA, Li Y, Nyquist MD, Miao L, Bradner JE, et al. Targeting chromatin binding regulation of constitutively active AR variants to overcome prostate cancer resistance to endocrine-based therapies. Nucleic Acids Res 2015; 43:5880-97.
|
| [41] |
McNair C, Urbanucci A, Comstock CE, Augello MA, Goodwin JF, Launchbury R, et al. Cell cycle-coupled expansion of AR activity promotes cancer progression. Oncogene 2017; 36:1655-68.
|
| [42] |
Luo J, Attard G, Balk SP, Bevan C, Burnstein K, Cato L, et al. Role of androgen receptor variants in prostate cancer: report from the 2017 mission androgen receptor variants meeting. Eur Urol 2018; 73:715-23.
|
| [43] |
Pomerantz MM, Li F, Takeda DY, Lenci R, Chonkar A, Chabot M, et al. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat Genet 2015; 47:1346-51.
|
| [44] |
Sharma NL, Massie CE, Ramos-Montoya A, Zecchini V, Scott HE, Lamb AD, et al. The androgen receptor induces a distinct transcriptional program in castration-resistant prostate cancer in man. Cancer Cell 2013; 23:35-47.
|
| [45] |
Stelloo S, Nevedomskaya E, Kim Y, Schuurman K, Valle- Encinas E, Lobo J, et al. Integrative epigenetic taxonomy of primary prostate cancer. Nat Commun 2018; 9:4900. https://doi.org/10.1038/s41467-018-07270-2.
pmid: 30464211
|
| [46] |
Bu H, Narisu N, Schlick B, Rainer J, Manke T, Schafer G, et al. Putative prostate cancer risk SNP in an androgen receptorbinding site of the melanophilin gene illustrates enrichment of risk SNPs in androgen receptor target sites. Hum Mutat 2016; 37:52-64.
|
| [47] |
Dadaev T, Saunders EJ, Newcombe PJ, Anokian E, Leongamornlert DA, Brook MN, et al. Fine-mapping of prostate cancer susceptibility loci in a large meta-analysis identifies candidate causal variants. Nat Commun 2018; 9:2256. https://doi.org/10.1038/s41467-018-04109-8.
doi: 10.1038/s41467-018-04109-8
pmid: 29892050
|
| [48] |
Marshall TW, Link KA, Petre-Draviam CE, Knudsen KE. Differential requirement of SWI/SNF for androgen receptor activity. J Biol Chem 2003; 278:30605-13.
|
| [49] |
Nair SS, Guo Z, Mueller JM, Koochekpour S, Qiu Y, Tekmal RR, et al. Proline-, glutamic acid-, and leucine-rich protein- 1/modulator of nongenomic activity of estrogen receptor enhances androgen receptor functions through LIM-only coactivator, four-and-a-half LIM-only protein 2. Mol Endocrinol 2007; 21:613-24.
|
| [50] |
Mills IG, Gaughan L, Robson C, Ross T, McCracken S, Kelly J, et al. Huntingtin interacting protein 1 modulates the transcriptional activity of nuclear hormone receptors. J Cell Biol 2005; 170:191-200.
|
| [51] |
Gao S, Wang H, Lee P, Melamed J, Li CX, Zhang F, et al. Androgen receptor and prostate apoptosis response factor-4 target the c-FLIP gene to determine survival and apoptosis in the prostate gland. J Mol Endocrinol 2006; 36:463-83.
|
| [52] |
Liu S, Kumari S, Hu Q, Senapati D, Venkadakrishnan VB, Wang D, et al. A comprehensive analysis of coregulator recruitment, androgen receptor function and gene expression in prostate cancer. Elife 2017; 6:e28482. https://doi.org/10.7554/eLife.28482.
doi: 10.7554/eLife.28482
pmid: 28826481
|
| [53] |
Jeter CR, Liu B, Lu Y, Chao HP, Zhang D, Liu X, et al. NANOG reprograms prostate cancer cells to castration resistance via dynamically repressing and engaging the AR/FOXA1 signaling axis. Cell Discov 2016;2:16041. https://doi.org/10.1038/celldisc.2016.41.eCollection2016.
pmid: 27867534
|
| [54] |
Culig Z. Androgen receptor coactivators in regulation of growth and differentiation in prostate cancer. J Cell Physiol 2016; 231:270-4.
|
| [55] |
Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010; 18:11-22.
|
| [56] |
Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet 2012; 44:685-9.
|
| [57] |
Imperato-McGinley J, Zhu YS. Androgens and male physiology the syndrome of 5alpha-reductase-2 deficiency. Mol Cell Endocrinol 2002; 198:51-9.
|
| [58] |
Taplin ME, Bubley GJ, Shuster TD, Frantz ME, Spooner AE, Ogata GK, et al. Mutation of the androgen-receptor gene in metastatic androgen-independent prostate cancer. N Engl J Med 1995; 332:1393-8.
|
| [59] |
Helsen C, Claessens F. Looking at nuclear receptors from a new angle. Mol Cell Endocrinol 2014; 382:97-106.
|
| [60] |
Norris JD, Joseph JD, Sherk AB, Juzumiene D, Turnbull PS, Rafferty SW, et al. Differential presentation of protein interaction surfaces on the androgen receptor defines the pharmacological actions of bound ligands. Chem Biol 2009; 16:452-60.
|
| [61] |
Kazmin D, Prytkova T, Cook CE, Wolfinger R, Chu TM, Beratan D, et al. Linking ligand-induced alterations in androgen receptor structure to differential gene expression: a first step in the rational design of selective androgen receptor modulators. Mol Endocrinol 2006; 20:1201-17.
|
| [62] |
Verrijdt G, Schoenmakers E, Haelens A, Peeters B, Verhoeven G, Rombauts W, et al. Change of specificity mutations in androgen-selective enhancers. Evidence for a role of differential DNA binding by the androgen receptor. J Biol Chem 2000; 275:12298-305.
|
| [63] |
Reid KJ, Hendy SC, Saito J, Sorensen P, Nelson CC. Two classes of androgen receptor elements mediate cooperativity through allosteric interactions. J Biol Chem 2001; 276:2943-52.
|
| [64] |
Hsiao PW, Thin TH, Lin DL, Chang C. Differential regulation of testosterone vs. 5alpha-dihydrotestosterone by selective androgen response elements. Mol Cell Biochem 2000; 206:169-75.
|
| [65] |
Gottlieb B, Beitel LK, Nadarajah A, Paliouras M, Trifiro M. The androgen receptor gene mutations database: 2012 update. Hum Mutat 2012; 33:887-94.
|
| [66] |
Paltoglou S, Das R, Townley SL, Hickey TE, Tarulli GA, Coutinho I, et al. Novel androgen receptor coregulator GRHL2 exerts both oncogenic and antimetastatic functions in prostate cancer. Cancer Res 2017; 77:3417-30.
|
| [67] |
Groner AC, Cato L, de Tribolet-Hardy J, Bernasocchi T, Janouskova H, Melchers D, et al. TRIM24 is an oncogenic transcriptional activator in prostate cancer. Cancer Cell 2016; 29:846-58.
|
| [68] |
Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 2012; 338:1465-9.
|
| [69] |
Sharma NL, Massie CE, Butter F, Mann M, Bon H, Ramos- Montoya A, et al. The ETS family member GABPalpha modulates androgen receptor signalling and mediates an aggressive phenotype in prostate cancer. Nucleic Acids Res 2014; 42:6256-69.
|
| [70] |
Masiello D, Cheng S, Bubley GJ, Lu ML, Balk SP. Bicalutamide functions as an androgen receptor antagonist by assembly of a transcriptionally inactive receptor. J Biol Chem 2002; 277:26321-6.
|
| [71] |
Kang Z, Janne OA, Palvimo JJ. Coregulator recruitment and histone modifications in transcriptional regulation by the androgen receptor. Mol Endocrinol 2004; 18:2633-48.
|
| [72] |
Baek SH, Ohgi KA, Nelson CA, Welsbie D, Chen C, Sawyers CL, et al. Ligand-specific allosteric regulation of coactivator functions of androgen receptor in prostate cancer cells. Proc Natl Acad Sci U S A 2006; 103:3100-5.
|
| [73] |
van Royen ME, van Cappellen WA, de Vos C, Houtsmuller AB, Trapman J. Stepwise androgen receptor dimerization. J Cell Sci 2012; 125:1970-9.
|
| [74] |
Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 2011; 474:390-4.
|
| [75] |
Jehle K, Cato L, Neeb A, Muhle-Goll C, Jung N, Smith EW, et al. Coregulator control of androgen receptor action by a novel nuclear receptor-binding motif. J Biol Chem 2014; 289:8839-51.
|
| [76] |
Guseva NV, Rokhlin OW, Bair TB, Glover RB, Cohen MB. Inhibition of p53 expression modifies the specificity of chromatin binding by the androgen receptor. Oncotarget 2012; 3:183-94.
|
| [77] |
Geserick C, Meyer HA, Haendler B. The role of DNA response elements as allosteric modulators of steroid receptor function. Mol Cell Endocrinol 2005; 236:1-7.
|
| [78] |
Buzon V, Carbo LR, Estruch SB, Fletterick RJ, Estebanez- Perpina E. A conserved surface on the ligand binding domain of nuclear receptors for allosteric control. Mol Cell Endocrinol 2012; 348:394-402.
|
| [79] |
Kumar R. Steroid hormone receptors and prostate cancer: role of structural dynamics in therapeutic targeting. Asian J Androl 2016; 18:682-6.
|
| [80] |
Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science 2009; 324:407-10.
|
| [81] |
Margeat E, Bourdoncle A, Margueron R, Poujol N, Cavailles V, Royer C. Ligands differentially modulate the protein interactions of the human estrogen receptors alpha and beta. J Mol Biol 2003; 326:77-92.
|
| [82] |
Deegan BJ, Bhat V, Seldeen KL, McDonald CB, Farooq A. Genetic variations within the ERE motif modulate plasticity and energetics of binding of DNA to the ERalpha nuclear receptor. Arch Biochem Biophys 2011; 507:262-70.
|
| [83] |
Geserick C, Meyer HA, Barbulescu K, Haendler B. Differential modulation of androgen receptor action by deoxyribonucleic acid response elements. Mol Endocrinol 2003; 17:1738-50.
|
| [84] |
Callewaert L, Verrijdt G, Haelens A, Claessens F. Differential effect of small ubiquitin-like modifier (SUMO)-ylation of the androgen receptor in the control of cooperativity on selective versus canonical response elements. Mol Endocrinol 2004; 18:1438-49.
|
| [85] |
Wei L, Wang J, Lampert E, Schlanger S, DePriest AD, Hu Q, et al. Intratumoral and intertumoral genomic heterogeneity of multifocal localized prostate cancer impacts molecular classifications and genomic prognosticators. Eur Urol 2017; 71:183-92.
|
| [86] |
Geng C, He B, Xu L, Barbieri CE, Eedunuri VK, Chew SA, et al. Prostate cancer-associated mutations in speckle-type POZ protein (SPOP) regulate steroid receptor coactivator 3 protein turnover. Proc Natl Acad Sci U S A 2013; 110:6997-7002.
|
| [87] |
Antonelli A, Palumbo C, Veccia A, Grisanti S, Triggiani L, Zamboni S, et al. Biological effect of neoadjuvant androgendeprivation therapy assessed on specimens from radical prostatectomy: a systematic review. Minerva Urol Nefrol 2018; 70:370-9.
|
| [1] |
Renee E. Vickman,Omar E. Franco,Daniel C. Moline,Donald J. Vander Griend,Praveen Thumbikat,Simon W. Hayward. The role of the androgen receptor in prostate development and benign prostatic hyperplasia: A review[J]. Asian Journal of Urology, 2020, 7(3): 191-202. |
| [2] |
Simeng Wen,Yuanjie Niu,Haojie Huang. Posttranslational regulation of androgen dependent and independent androgen receptor activities in prostate cancer[J]. Asian Journal of Urology, 2020, 7(3): 203-218. |
| [3] |
Ieva Eringyte,Joanna N. Zamarbide Losada,Sue M. Powell,Charlotte L. Bevan,Claire E. Fletcher. Coordinated AR and microRNA regulation in prostate cancer[J]. Asian Journal of Urology, 2020, 7(3): 233-250. |
| [4] |
Yezi Zhu,Jun Luo. Regulation of androgen receptor variants in prostate cancer[J]. Asian Journal of Urology, 2020, 7(3): 251-257. |
| [5] |
Chui Yan Mah,Zeyad D. Nassar,Johannes V. Swinnen,Lisa M. Butler. Lipogenic effects of androgen signaling in normal and malignant prostate[J]. Asian Journal of Urology, 2020, 7(3): 258-270. |
| [6] |
Ramesh Narayanan. Therapeutic targeting of the androgen receptor (AR) and AR variants in prostate cancer[J]. Asian Journal of Urology, 2020, 7(3): 271-283. |
| [7] |
Abhishek Tripathi,Shilpa Gupta. Androgen receptor in bladder cancer: A promising therapeutic target[J]. Asian Journal of Urology, 2020, 7(3): 284-290. |
| [8] |
Michel Bolla,Ann Henry,Malcom Mason,Thomas Wiegel. The role of radiotherapy in localised and locally advanced prostate cancer[J]. Asian Journal of Urology, 2019, 6(2): 153-161. |
| [9] |
Pei Zhao,Yezi Zhu,Liang Cheng,Jun Luo. Detection of androgen receptor (AR) and AR-V7 in small cell prostate carcinoma: Diagnostic and therapeutic implications[J]. Asian Journal of Urology, 2019, 6(1): 109-113. |
| [10] |
Cameron M. Armstrong,Allen C. Gao. Current strategies for targeting the activity of androgen receptor variants[J]. Asian Journal of Urology, 2019, 6(1): 42-49. |
| [11] |
Michael V. Fiandalo,Daniel T. Gewirth,James L. Mohler. Potential impact of combined inhibition of 3α-oxidoreductases and 5α-reductases on prostate cancer[J]. Asian Journal of Urology, 2019, 6(1): 50-56. |
| [12] |
Tyler Etheridge,Shivashankar Damodaran,Adam Schultz,Kyle A. Richards,Joseph Gawdzik,Bing Yang,Vincent Cryns,David F. Jarrard. Combination therapy with androgen deprivation for hormone sensitive prostate cancer: A new frontier[J]. Asian Journal of Urology, 2019, 6(1): 57-64. |
| [13] |
Haley Dicken,Patrick J. Hensley,Natasha Kyprianou. Prostate tumor neuroendocrine differentiation via EMT: The road less traveled[J]. Asian Journal of Urology, 2019, 6(1): 82-90. |
| [14] |
Lingfan Xu,Junyi Chen,Weipeng Liu,Chaozhao Liang,Hailiang Hu,Jiaoti Huang. Targeting androgen receptor-independent pathways in therapy-resistant prostate cancer[J]. Asian Journal of Urology, 2019, 6(1): 91-98. |
| [15] |
Per-Anders Abrahamsson. Intermittent androgen deprivation therapy in patients with prostate cancer:Connecting the dots[J]. Asian Journal of Urology, 2017, 4(4): 208-222. |
|
|
|
|




